Andras Gyorgy

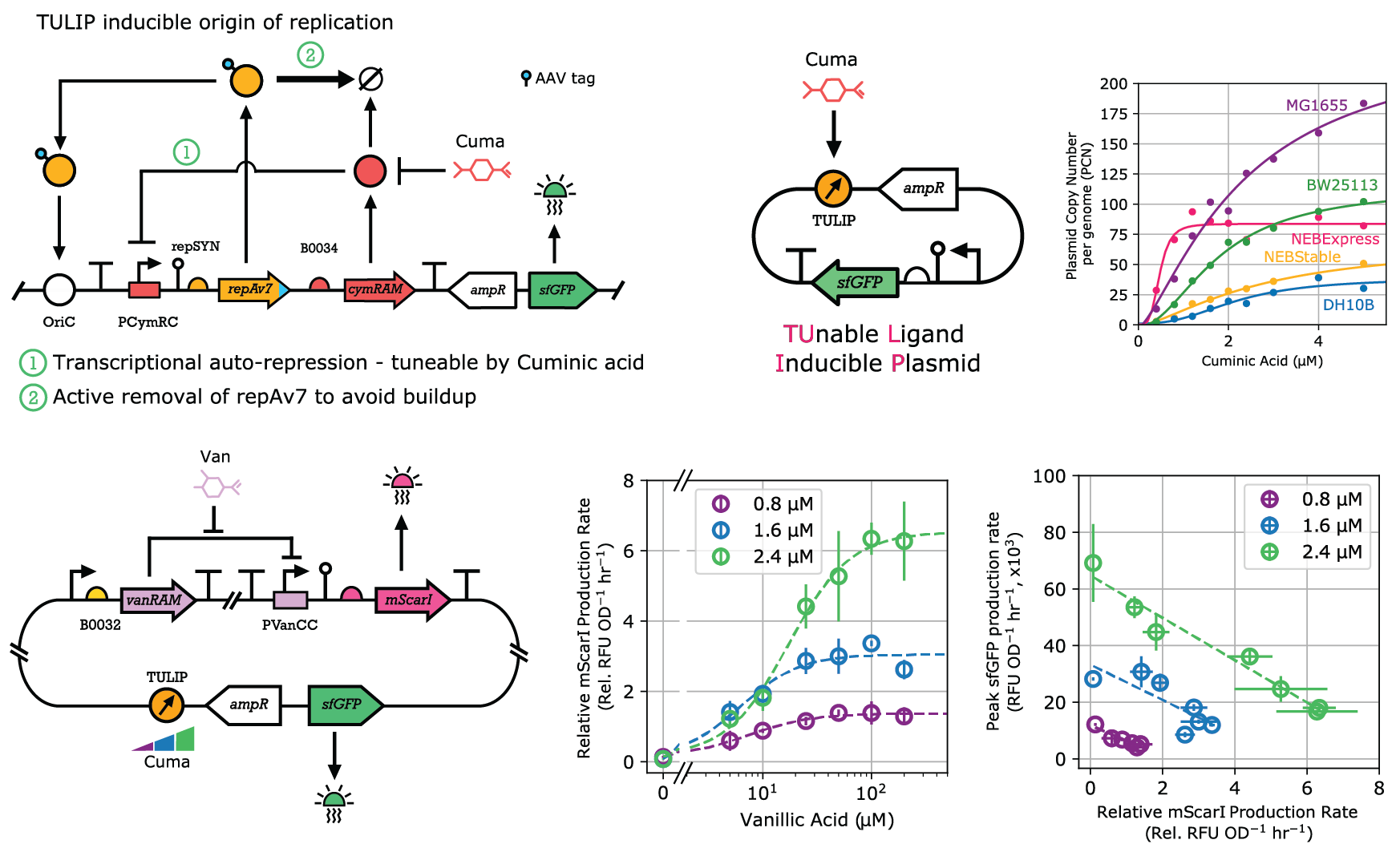
The ability to externally control gene expression has revolutionized biological research, particularly in synthetic biology. Such control typically occurs at the transcriptional and translational level, while technologies enabling control at the DNA copy level are limited by either (i) relying on a handful of plasmids with fixed and arbitrary copy numbers; or (ii) require multiple plasmids for replication control; or (iii) are restricted to specialized strains. To overcome these limitations, we developed TULIP (TUnable Ligand Inducible Plasmid): a portable plasmid with inducible copy number control, suitable for various Escherichia coli strains used in cloning, protein expression, and metabolic engineering. We demonstrated that flexible PCN control accelerates gene circuit design, optimizes metabolic burden probing, and facilitates module prototyping in different genetic contexts. We are currently working on expanding copy number control to other organisms and on leveraging it in a variety of synthetic biology applications. These include revealing the role of antibiotic selection markers on cellular performance, accelerating the loss of unwanted plasmids, enhancing modular cloning frameworks for rapid prototyping, and realizing intercellular communication via DNA messages. [more]
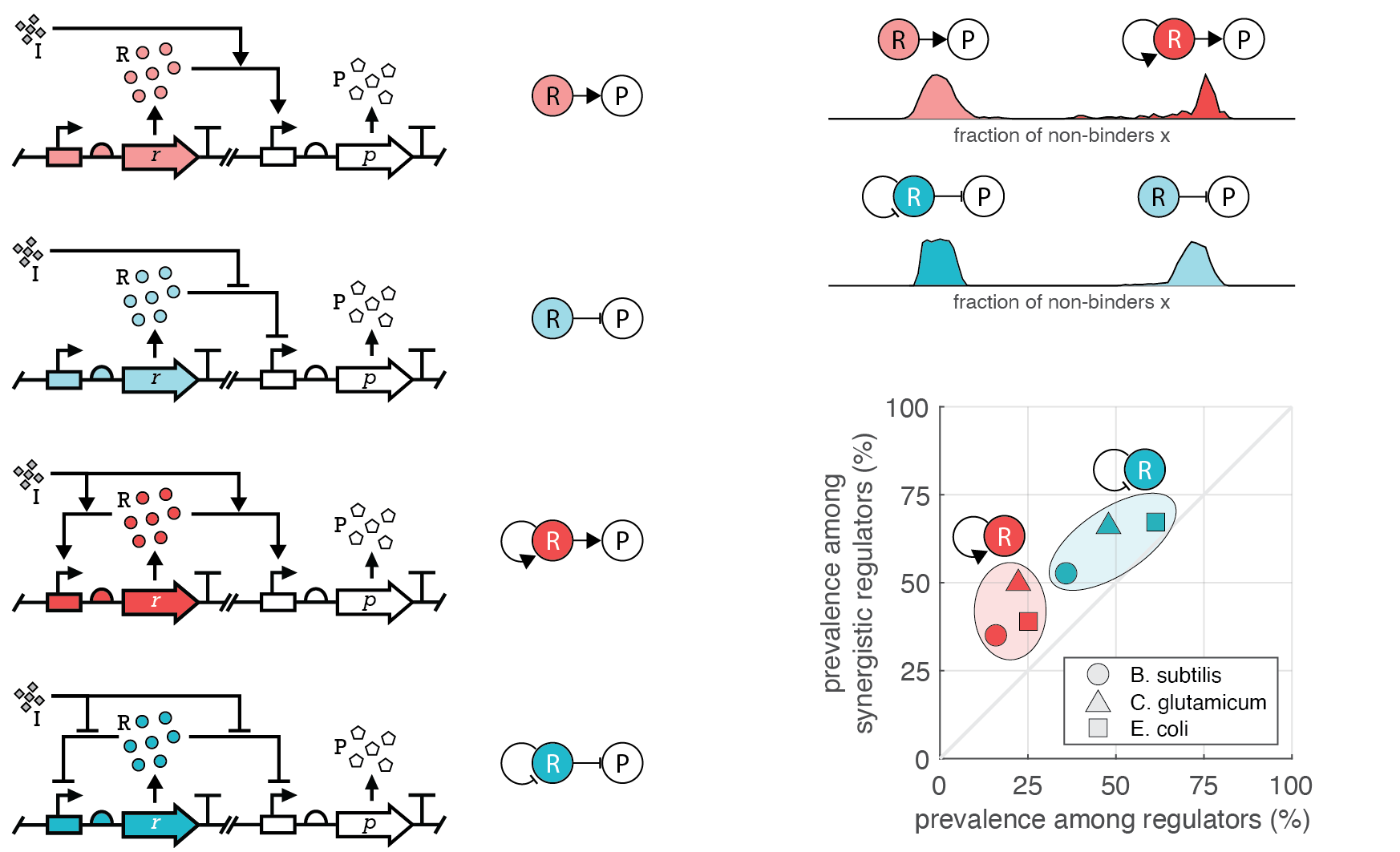
Gene products that are beneficial in one environment may become burdensome in another, prompting the emergence of diverse regulatory schemes that carry their own bioenergetic cost. By ensuring that regulators are only expressed when needed, we demonstrated that autoregulation generally offers an advantage in an environment combining mutation and time-varying selection. Whether positive or negative feedback emerges as dominant depends primarily on the demand for the target gene product, typically to ensure that the detrimental impact of inevitable mutations is minimized. While self-repression of the regulator curbs the spread of these loss-of-function mutations, self-activation instead facilitates their propagation. By analyzing the transcription network of multiple model organisms, we revealed that reduced bioenergetic cost may contribute to the preferential selection of autoregulation among transcription factors. Inspired by these insights, we are currently incorporating evolutionary considerations into the the design and analysis of genetic modules. For instance, combining computational modeling and in vivo mutagenesis experiments in E. coli, we characterize how the interplay of gene dosage via plasmid copy number and regulatory architecture affect the phenotypic mutation rate, and how it depends on the type of mutations (gain-of-function vs. loss-of-function) and on the location of the mutation (coding region vs. regulatory region). [more]
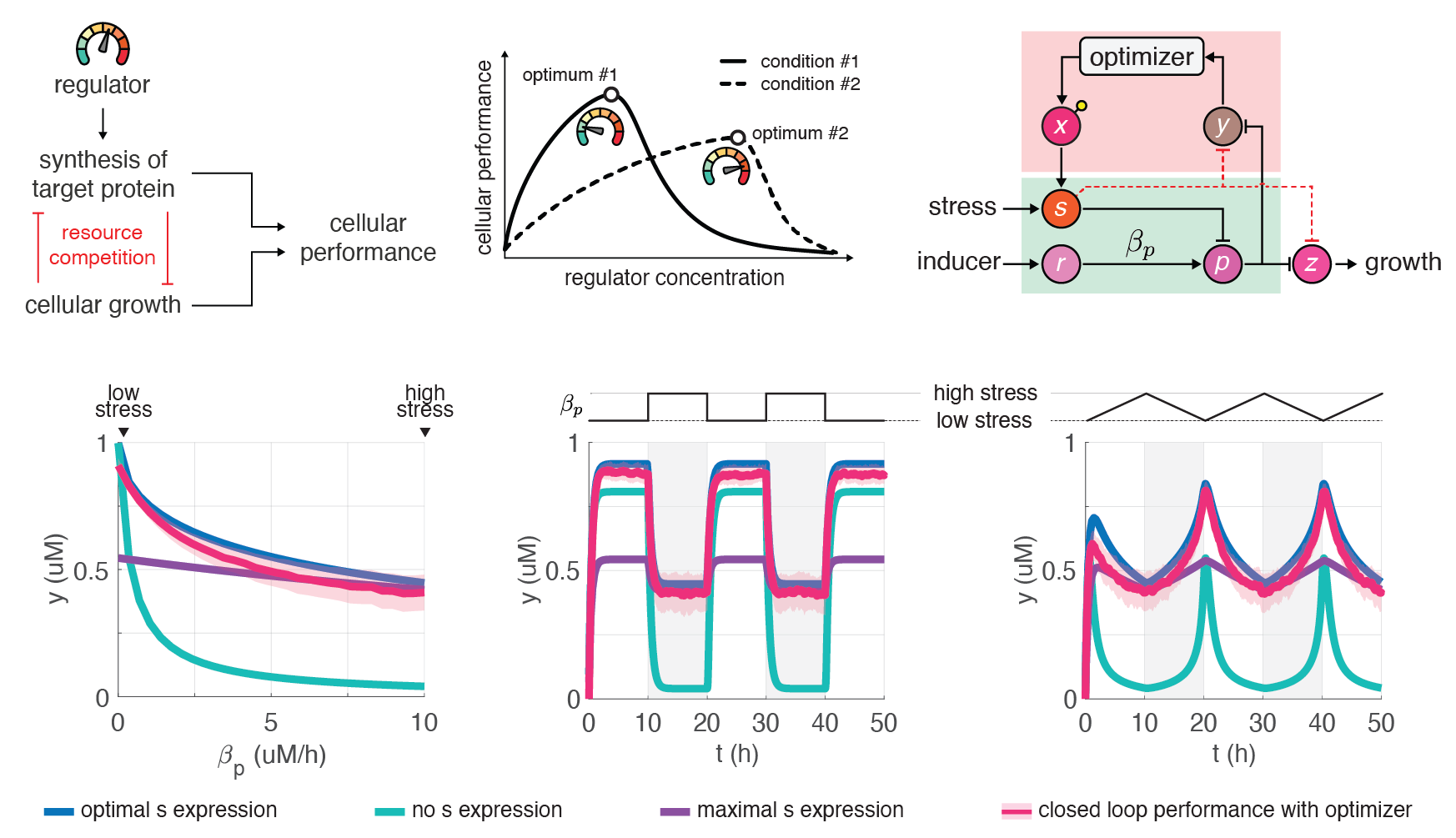
Biomolecular control enables leveraging cells as biomanufacturing factories. Despite recent advancements, we currently lack genetically encoded modules that can be deployed to dynamically fine-tune and optimize cellular performance. To address this shortcoming, we developed the blueprint of a genetic feedback module to optimize a broadly defined performance metric by adjusting the production and decay rate of a (set of) regulator species. We demonstrated that the optimizer can be implemented by combining available synthetic biology parts and components, and that it can be readily integrated with existing pathways and genetically encoded biosensors to ensure its successful deployment in a variety of settings. We further illustrated that the optimizer successfully locates and tracks the optimum in diverse contexts when relying on mass action kinetics-based dynamics and parameter values typical in E. coli. Since multistable switches represent key components in this optimizer module in addition to being core building blocks in natural systems, we now focus on understanding how their fundamental properties depend not only on the switches themselves, but also on their genetic context, for instance, due to the limited availability of shared cellular resources. [more]